On April 17th, 2024, a research article was published in Nature entitled "Digital colloid-enhanced Raman spectroscopy by single-molecule counting." Designed to overcome the long-standing challenge of quantitative reproducibility in the field of surface-enhanced Raman spectroscopy (SERS), digital colloid-enhanced Raman spectroscopy (dCERS) was developed based on the principle of single-molecule counting, achieving reproducible quantitative detection of target molecules at ultra-low concentrations and laying the critical foundation for the broad application of SERS.

Background
Raman spectra were discovered by Chandrasekhara Venkata Raman in 1928, who was awarded the 1930 Nobel Prize in Physics. Owing to inherently unique vibrational characteristics of molecules, this spectroscopy provides a “fingerprint” of individual molecular species, which thereby can be used for their identification. As no external labels are needed, this method has since found wide application in physics, chemistry, biology, geology, medicine, national defense and public security.
However, Raman signals are intrinsically weak, and so signal enhancement is crucial. Surface-enhanced Raman spectroscopy (SERS) was first discovered in 1974 by Martin Fleischmann at the University of Southampton. The phenomenon was explained in 1977 from an electromagnetic perspective by David L. Jeanmaire and Richard P. Van Duyne from Northwestern University and as owing to a charge-transfer effect by M. Grant Albrecht and J. Alan Creighton from the University of Kent. The surface enhancement of the signal can be so significant that single molecules can be detected with SERS, and so has been considered as possibly worthy of a second Nobel Prize. This area is indeed a vigorous area of research, as substrates with various surface morphologies have been developed with the goal of even higher enhancements, including nanostars, nanourchins, nanoflowers, nanoforests, among others. Nanostructures like tips and gaps were specifically created through wet chemistry and chip fabrication methods to produce intense electromagnetic hotspots to enable the detection of molecules at ultra-low concentrations.
However, although the field has continued to grow, there has emerged a persistent problem that has plagued all research: the signal intensities at low concentrations have been significantly irreproducible. Thus, while single molecules can be detected, accurate quantification of solution concentrations has heretofore not been possible. Thus, obtaining a platform that satisfies the dual requirements of high signal enhancement and reproducible quantification has remained a long-standing goal within the SERS community. Indeed, this issue has remained a major bottleneck that cannot be solved with any currently known technical frameworks.

Figure 1. Scheme for digital colloid-enhanced Raman spectroscopy.
dCERS achieves a quantitative sensitivity of less than 1 fM
The team of Professors Jian Ye and Zhifeng Shao from the School of Biomedical Engineering of SJTU developed digital colloid-enhanced Raman spectroscopy (dCERS). In this method, using colloidal nanoparticles, single molecules can be detected efficiently, even with significant differences in the intensities measured from different single molecules, which has been described in previous work. However, the difference is, in this method, the presence or absence of individual molecules is measured, pixel-by-pixel, based on the presence or absence of the specific Raman spectra of the target molecule. The number of positive detections is then simply counted, from which the concentration is determined.
This method can be used for the detection of a variety of molecules (such as dyes, metabolites, nucleic acids and proteins), with the limit of detection at least down to the fM (10-15 M) level (Figure 2). The synthesis of the SERS colloids is simple and can be easily scaled-up for larger-scale studies. In practice, a small amount from each batch of colloids is used to generate a calibration curve for the target molecule, which is then used to determine the concentration of the target in the quantitative analysis of samples in which the concentration is unknown.
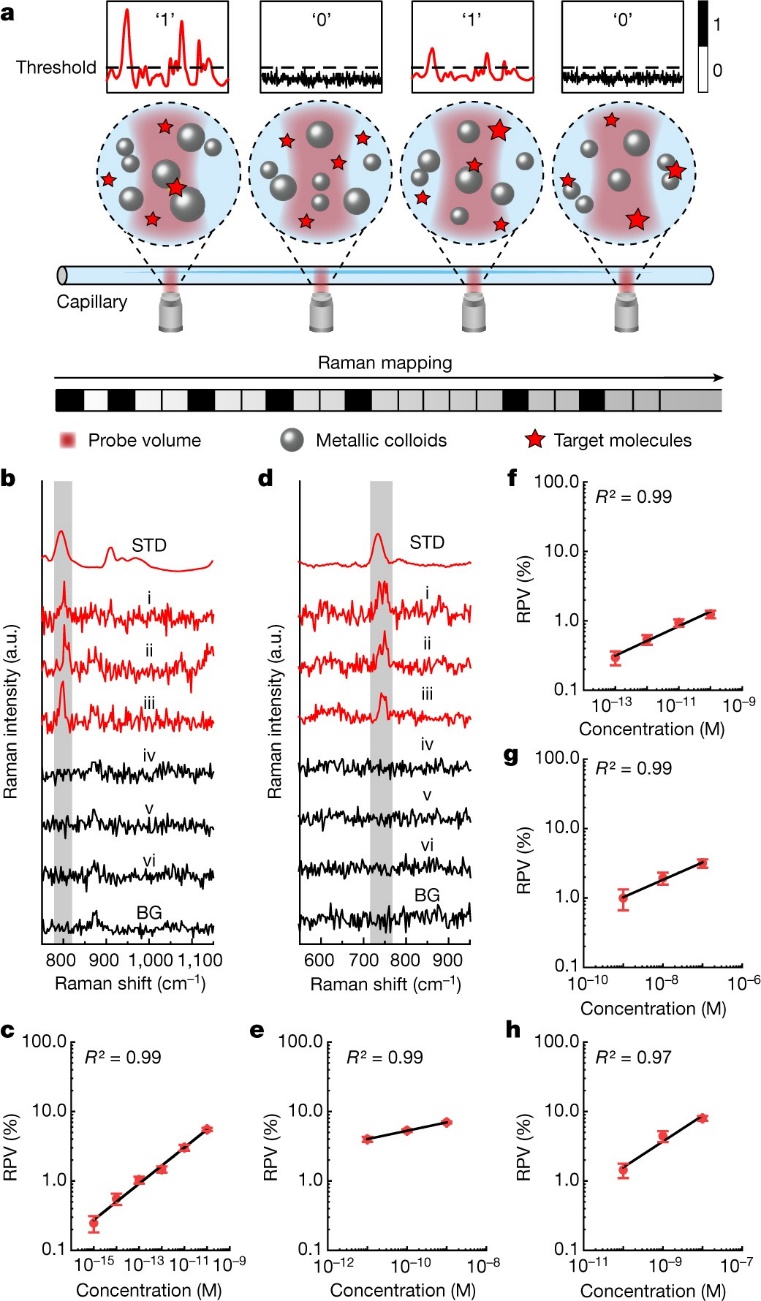
Figure 2. Principle of dCERS and the calibration curves of different molecules.
Quantitative reproducibility is highly controllable
In this work, the authors have found that the occurrence of the single-molecule events follows Poisson statistics. Thus, the quantitative sensitivity and accuracy can be determined simply by the number of positive detections, which is different from the conventional analog methods. As shown in Figure 3, by increasing the total number of spectra analyzed, there is an increase in the number of positive spectra, which effectively enhances the accuracy of the quantification. Thus, in real-world applications, the quantitative accuracy can be controlled according to the number of positive spectra detected (essentially, the total measurement time), based on the detection efficiency of the target molecules. Hence, dCERS is accurate, controllable, and reproducible.
The authors have also found that for different target molecules, although the detection efficiency may vary (thus, the requirement for pre-calibration), the overall adsorption behavior is well described by classical Gibbs thermodynamics. In fact, this is the first time that the physical mechanism underlying single-molecule counting has been established, which may be applicable to other single-molecule counting-based techniques besides dCERS.

Figure 3. dCERS quantitative error follows Poisson statistics.
Applications for Environmental Protection and Food Safety
To further explore the potential real-world applications of dCERS, two highly toxic molecules, paraquat and thiram, were examined in further detail (as shown in Figure 4). Paraquat is an extremely toxic herbicide that may cause Parkinson's disease and is banned in 32 countries. Thiram is a sulfur-containing, toxic fungicide classified as a category 2 carcinogen by the European Union. Thus, a highly sensitive, accurate, and reliable quantitative detection technology is particularly important for detecting such toxic molecules. In principle, there may be no so-called “safe dose” for such carcinogens.
In this study, normal lake water was used as the background and mixed with trace amount of paraquat. A detection sensitivity for paraquat that is three orders of magnitude lower than the maximum residue levels specified by the European Union was achieved. As for thiram, this molecule was added to lab-grown bean sprout extracts and a detection sensitivity that is orders of magnitude better than what can be obtained with mass spectrometry was achieved. It was demonstrated that after a series of dilutions, interference from other molecules in the samples could be effectively reduced, which thereby enabled the accurate detection of each target molecule. It is worth mentioning that, the high sensitivity and reliable statistics of dCERS is the solid foundation for such quantification.
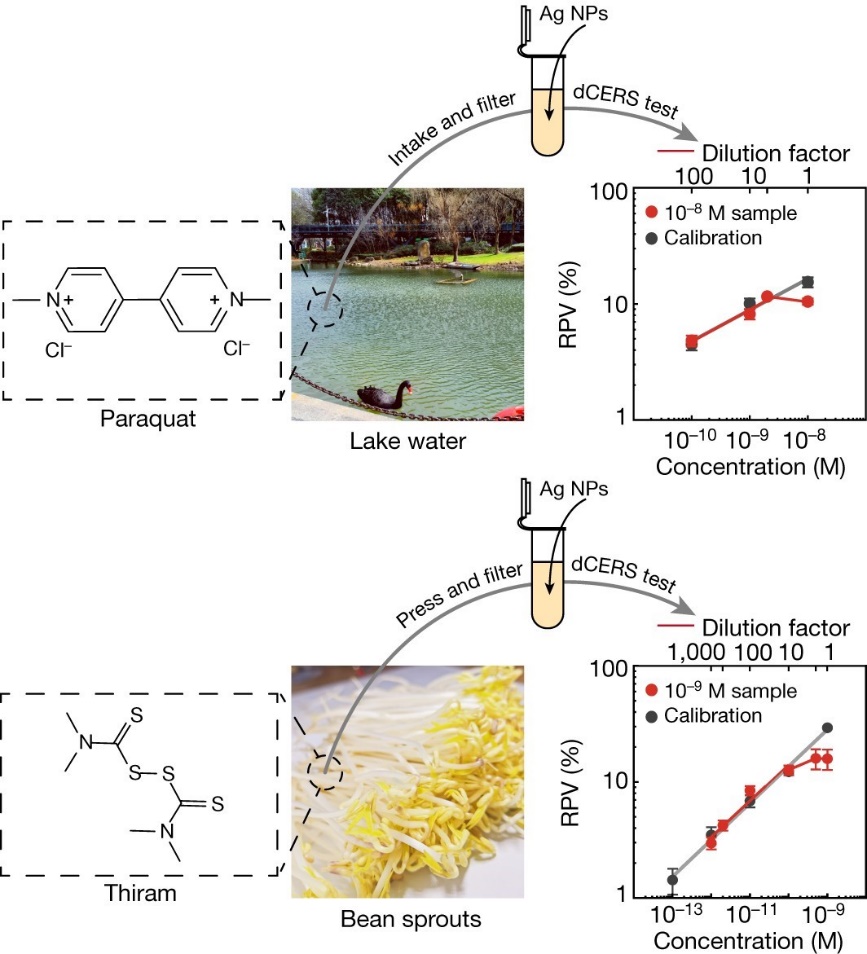
Figure 4. Quantitative detection of trace amount of chemicals by dCERS.
This work demonstrated that dCERS, based on single-molecule counting, can achieve reproducible quantification of ultra-low concentrations of target molecules in unknown backgrounds without any external labels. Since most molecules have distinct SERS spectral signatures, dCERS is suitable for the quantitative detection of various molecules even at the same time. Additionally, dCERS offers the straightforward ability to scale-up production with tractability and low cost as well as simple measurement procedures, which is expected to further promote the revolution and the progress of this highly sensitive detection technology.
As this year marks the 50th anniversary of the discovery of SERS, it is expected that the broad application of SERS will be promoted in fields such as life sciences, clinical medicine, environmental protection, food examination, national defense and public security, as the further development of dCERS.

Figure 5. Authors of this article. From left to right: Zhifeng Shao, Jian Ye, Xinyuan Bi, and Daniel M. Czajkowsky.
Website of Jian Ye’s group: http://www.yelab.sjtu.edu.cn
There are four authors of this publication, all from the School of Biomedical Engineering department at Shanghai Jiao Tong University. The first author is Xinyuan Bi, a Ph.D. candidate in the group of Professor Jian Ye, who is the corresponding author. The other senior author of this work is Professor Zhifeng Shao, who made key contributions in the basic concepts, data analysis, and writing of the article. The other author is Professor Daniel M. Czajkowsky, who made important contributions to the interpretation of the physical principles of the data and the revision of the article.
This work was assisted by Prof. Hongchen Gu, Prof. Hong Xu and Prof. Feng Shen from Shanghai Jiao Tong University, and supported by the National Natural Science Foundation of China, National Key R&D Program of China, Science and Technology Commission of Shanghai Municipality, Shanghai Key Laboratory of Gynecologic Oncology, and Shanghai Jiao Tong University, and the K. C. Wong Education Foundation (Hong Kong).
Link: https://www.nature.com/articles/s41586-024-07218-1
Editor on Duty: Yan Cheng

